Technology for single-cell analysis
- Shotgun single-cell proteomics
- Prioritized single-cell proteomics
- Parallel analysis of both single cells and peptides
- Sample preparation for single-cell proteomics
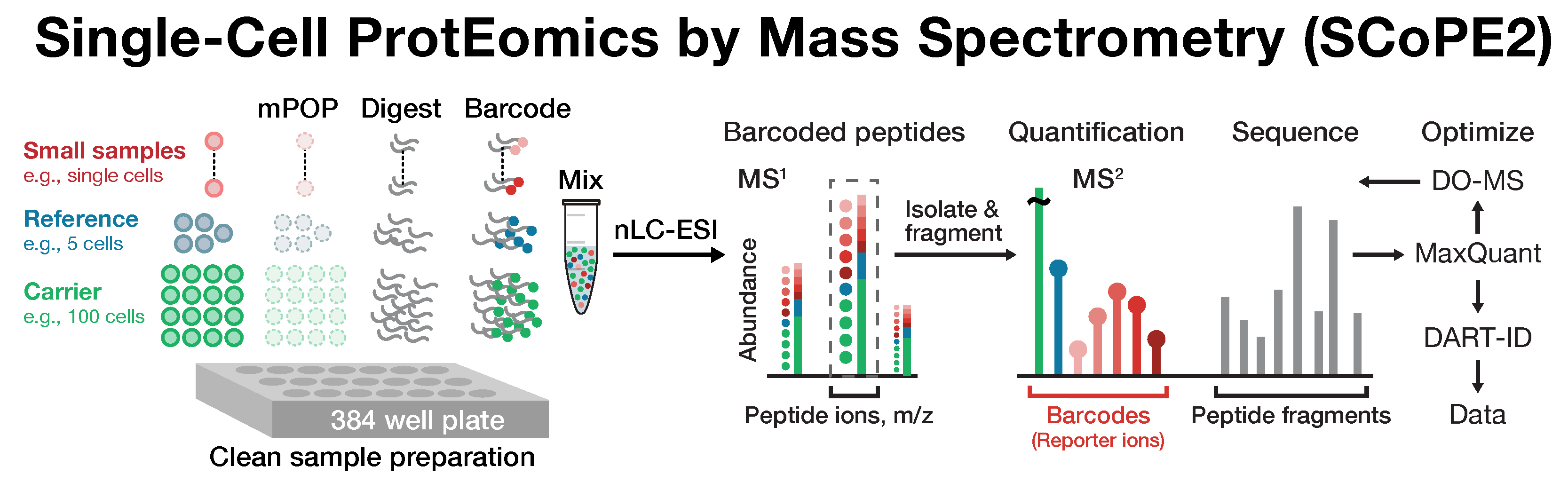
Shotgun single-cell proteomics
SCoPE-MS and its second version SCoPE2 can analyze thousands of proteins selected in order of their abundance in the samples. SCoPE2 uses multiplexed experimental designs in which proteins from single cells and from a small group of cells (called carrier proteins) are barcoded with isobaric mass tags and then combined. This design, utilizes the isobaric carrier to reduce the loss of proteins from single cells adhering to equipment surfaces while simultaneously enhancing peptide identification.
Prioritized single-cell proteomics
pSCoPE can analyze thousands of proteins selected in order of their priority for the biological question and project of interest. pSCoPE achieves high data completeness within, usually above 85%.
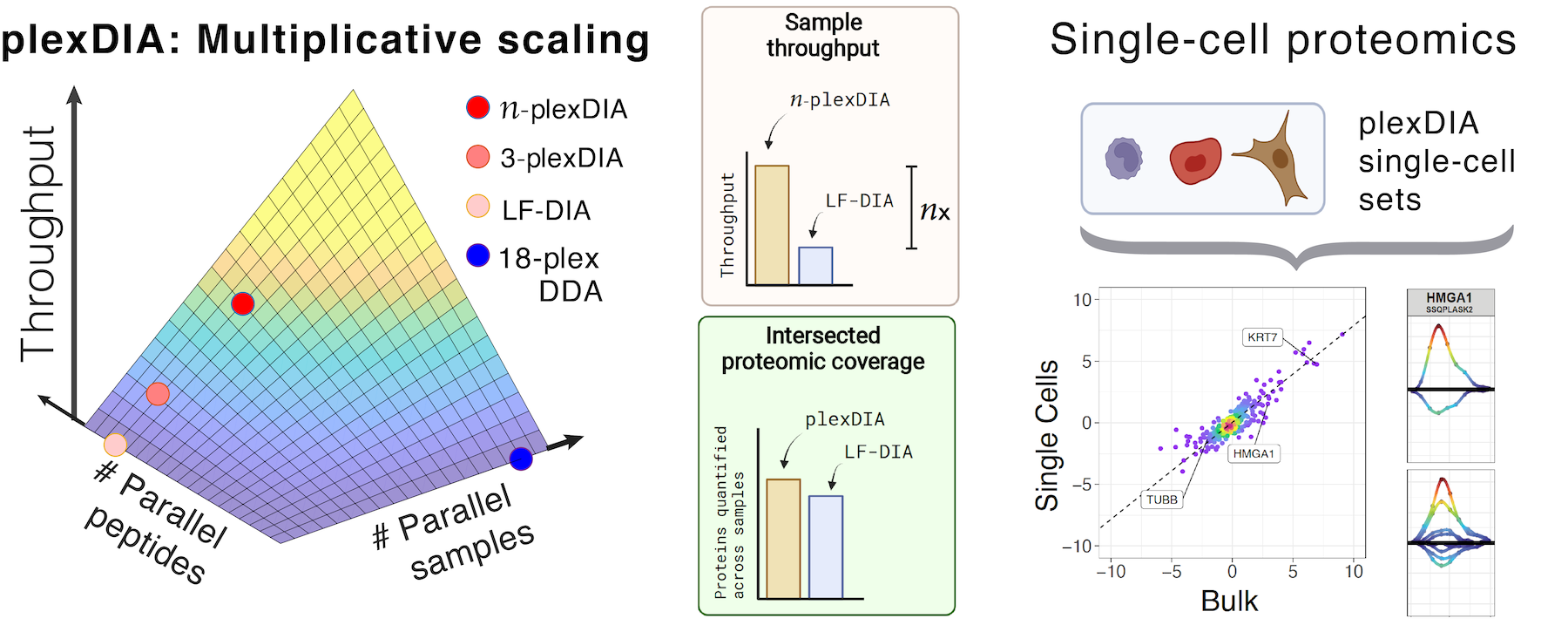
Parallel analysis of both single cells and peptides
plexDIA parallelizes the analysis of both single cells and peptides. It quantifies 1,000 - 5,000 proteins per single cell (depending on cell size and other factors) while achieving high data completeness within a set, usually above 90 %. It does not use a carrier sample, though it serves as a basis for SCoPE-DIA, which uses an isotopologous carrier sample.
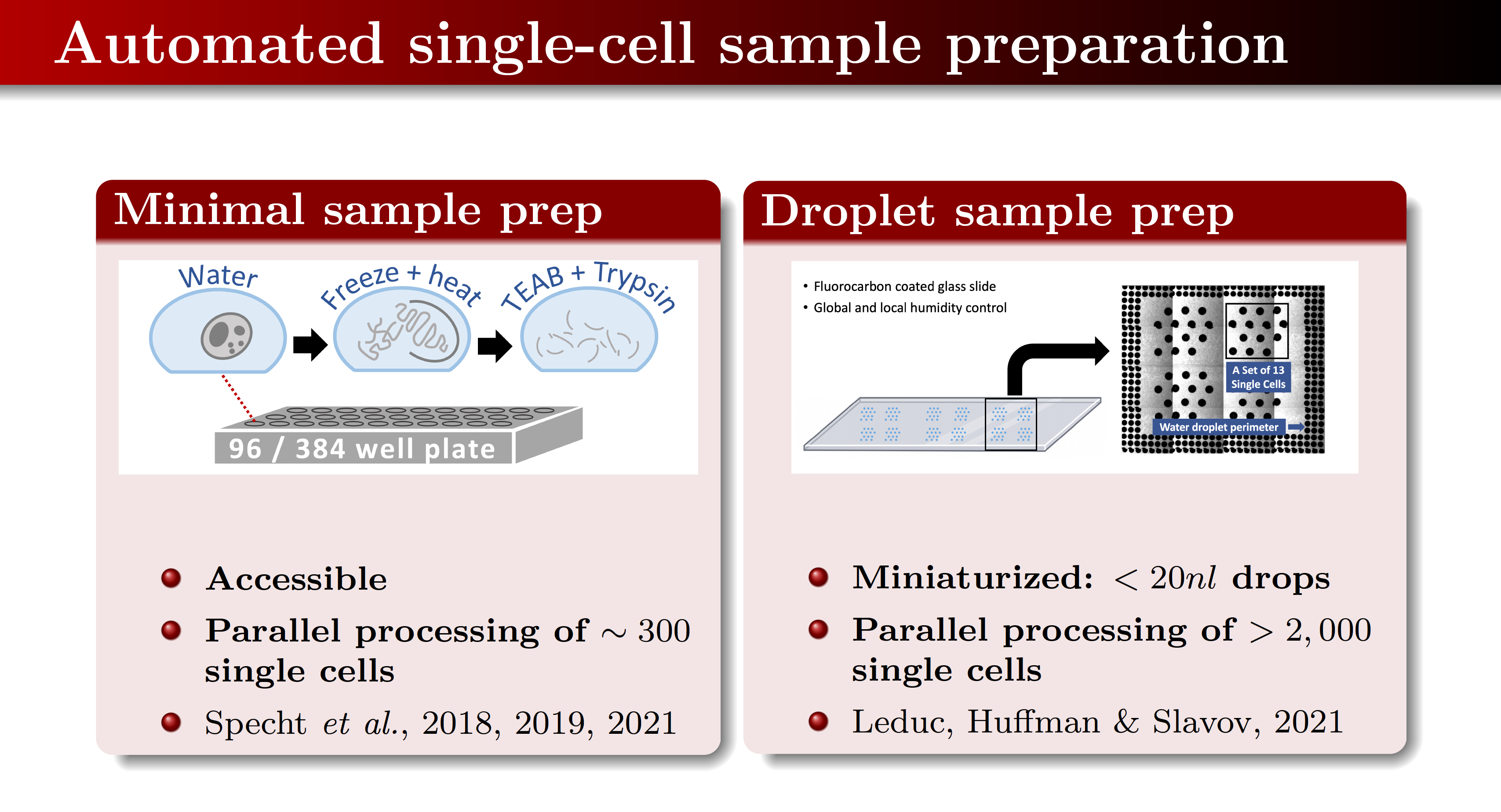
Sample preparation for single-cell proteomics
We have developed and use methods that use solely MS-compatible reagents and allow parallel preparation of hundreds of single cells in small volumes. These include minimal sample preparation and droplet sample preparation.
About the single-cell proteomics center
The single-cell proteomics center is located at Northeastern University in Boston, MA, USA. Questions about the center should be addressed to Prof. Nikolai Slavov.